Introduction
Mapped RNA-Seq data coming from eukaryotes is probably the most complicated data type to import into SeqMonk due to it’s relative complexity and the abundance of options with which you are presented. Depending on exactly what sort of information you want to know about your data different data import options will be useful, so understanding the best way to import your data can be important.
Options
Virtually all RNA-Seq data will start out as a BAM / SAM file since this is the only format currently capable of describing mapped positions which span one or more splice junctions. The options for import are therefore the options found in the BAM / SAM parser. The options you have are:
- Is the data single or paired end
- Do you want to split the reads into spliced segments
- Do you want to import introns instead of exons
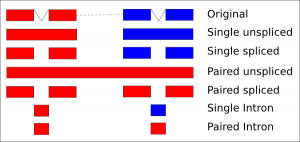
Each combination of options will produce a different set of aligned reads and will be useful for different types of analysis. The figure below summarises the reads which are imported from a set of spliced paired end mapped data. The colours represent alignment direction (red=forward, blue=reverse).
Choosing the correct options
The options you choose will depend on what you want to do with your data. Typically you either want to do one of three things:
- Quantitatively analyse the expression of each transcript
- Quantitatively analyse splice variation
- Identify novel transcripts
If you want to quantitatively analyse expression then you need to split your reads so that separate reads are recorded for each mapped splice section. You should then combine this with the base pair quantiation so that each spliced read is counted proportionally in the different exons it spans. If you have paired end data then selecting paired end import will simply reverse the orientation of any reads coming from the second read in a pair. If your data is not directional then this won’t make any difference, but if you have a directional library this will ensure that the correct orientation is assigned to each read.
If your interest is in the analysis of splice variants then the easiest way to do this is to import only the introns from your mapped data. Introns will only be imported from a read which contains an internal splice site, reads which do not splice will be ignored. Once the introns have been imported these should be quantitated using the read position probe generator, to put a probe over every different splice junction combination and then quantitated with the exact overlap quantitation to get an exact count for each splice combination seen in the data. If you have paired end data then it’s best to import introns as paired end data so that splice junctions from the second read match exactly (position and strand) with those from the first read.
Finally, if your interest is in seeing the extend of novel transcripts then you can import spliced mapped data as a normal BAM / SAM file. Since the mapped region is not expected to be contiguous on the genome you will need to greatly increase the filter for largest allowed distance between the ends of the reads to avoid rejecting reads which span introns. This type of import will join the ends of the read (or reads for paired end data) so you get the most complete view of the region of the genome spanned by your reads, but the quantitative influence of each read will depend on the size of the introns spanned, so importing data this way should only be used as a qualitative way of viewing your data. Combining this view of your transcriptome with a more conventional spliced import in a different track should give you the most complete view of the position and extent of each of your transcripts.