Introduction
After a long gestation we’ll be releasing a new version of FastQC in the near future to address some of the common problems and confusions we’ve encountered in the current version. I’ll write more about this in future posts but wanted to start with the most common complaint, that the duplicate sequence plot was difficult to understand and didn’t really show the information people wanted to see. I wrote a post previously to explain how the current module works but in the new release this module will be significantly restructured to give a more comprehensive, relevant and hopefully intuitive view of the duplication within a library.
What was wrong with the old version?
A few things actually…
- To keep memory usage sane we had to sample the library rather than count all sequences. This meant that what we showed in the plot was the relative number of sequences within the sample set with different duplication levels. Because there wasn’t enough space to show all possible duplication levels we had to set a limit at 10, but this meant that the plot couldn’t accurately reflect the level of duplication just by looking at the graph, because you couldn’t tell whether the sequences in the 10+ field occurred 11 or 11,000 times.
- Because we hadn’t found a nice way to extrapolate from the subset of data we monitored to the whole dataset, the y axis for the plot expressed duplication levels relative to unique sequences. This was easy to do but was difficult to interpret, and was pretty misleading in cases where there were virtually no unique sequences in the library.
- We included an overall duplication value at the top of the plot, which was a percentage value, but again this value was difficult to interpret since the value shown was simply the percentage of sequences in the library which were not unique. What this meant was that a library containing the same sequence in every read, and a library with diverse sequences which were all present exactly twice would both have a duplication level of 100%, which wasn’t very good.
What have we done to fix this?
The new version of the plot still retains some of the properties (and limitations) of the old method. Basically, the way in which the initial count data is collected hasn’t changed. We still sample the library so that we don’t require huge amounts of memory, and we still only look at the first 50bp of any library over 75bp so we don’t get messed up by base calling error. It’s the way we treat and represent this data which has changed.
The inspiration for the new version of the module mainly revolves around an improved way of extrapolating from our limited dataset to the rest of the library so that we can accurately predict what we would have seen if we had actually measured the whole library. The maths for this was kindly worked out and written up for us by Brian Howard of sciome.com, to whom we’re extremely grateful, and we’ve adapted his approach in the new version.
The new plot has three major changes to the old one.
- The number of x-axis categories has been increased to expand the 10+ category into several groups to give you a clearer picture of the true level of duplication in your sample.
- The y-axis of the plot now represents a percentage of the total library, so all values are directly comparable and much easier to understand.
- The plot now contains two lines instead of one. Both of these profiles show the proportion of the library which comes from sequences with different levels of duplication. The two traces simply show the percentages of the raw and deduplicated libraries. By looking at the two traces you can quickly see how the library is divided between different duplication levels, and by comparing the two traces you can easily see whether the 10+ category is composed of a small number of very highly duplicated sequences, or a much larger number of moderately duplicated sequences.
In addition to the changes to the plots we have also changed the headline duplication level value. This is still shown as a percentage, but the value now shows what percentage of the library would remain if you deduplicated it to keep only one copy of every different sequence, which fits a lot better with how people actually think about duplication.
So is it now perfect?
Yes.
OK, no. But it’s much better. There are however some problems which aren’t solved (and may not be solvable).
- The sampling of the library is still based on the first N different sequences we see (where N is currently 200,000 but in the new scheme it doesn’t really matter too much what it is). This approach relies of seeing the sequences in a random order which is fine for fastq files, but you can still get misleading answers if you feed a sorted BAM file into the program. This isn’t really something we can solve since the only guaranteed solution involves holding every possible sequence in memory, which is bad.
- When setting warn / error limits we have to make assumptions about the library which aren’t really true. We assume that we’re sampling from an infinitely sized genome, and that no duplication is always better. In reality you might be sampling very deeply from a small genome where you’d expect a certain level of duplication, and where lower duplication levels might be just as worrying as high levels. Alternatively you might be sampling in a way where even representation isn’t expected (such as RNA-seq libraries where representation reflects expression levels). In theory we could address the first case if we were given a genome size, but the second case is very difficult unless we make some big assumptions about the expected distribution of expression levels. This might be more of a problem if library sizes got much bigger, but it’s not too bad at the moment.
- All of the duplication detection is based on an exact match between two sequences and even a single base change is enough to call the sequences as different. In libraries of poor quality, which contain a larger percentage of sequencing errors, the duplication level will be under-reported as the sequencing errors will be incorrectly interpreted as diversity in the library. This might be theoretically solvable with fuzzy matching, or a quality aware algorithm, but both of these would be significantly more computationally intensive than the current approach and would slow down the whole program – something we’re trying very hard not to do.
Can I see an example?
Of course. Let’s start with a nice library and work our way down to something nasty.
Example 1 – Very diverse library
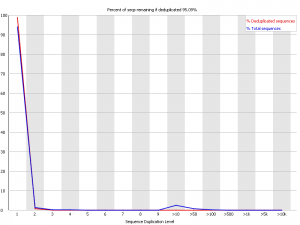 Here we have a library with low duplication. In both the raw and deduplicated versions of the library the vast majority of reads come from sequences which only occur once within the library. This suggests that the library sampling isn’t anywhere close to saturation and that you have a diverse population.
Here we have a library with low duplication. In both the raw and deduplicated versions of the library the vast majority of reads come from sequences which only occur once within the library. This suggests that the library sampling isn’t anywhere close to saturation and that you have a diverse population.
Example 2 – RNA-Seq
 This is a good RNA-Seq library, but actually represents the kind of profile you might see in any enriched experiment. In general FastQC makes the assumption that you expect to be looking at a diverse library with a roughly even representation of all sequences. In enriched libraries this isn’t going to be true and there are certain sequences (highly expressed genes or enriched peaks in a ChIP) where you want the sequence to occur more frequently. Above a certain read density the only way to place more reads into a region is via duplication so we might not expect to have a properly diverse library.
This is a good RNA-Seq library, but actually represents the kind of profile you might see in any enriched experiment. In general FastQC makes the assumption that you expect to be looking at a diverse library with a roughly even representation of all sequences. In enriched libraries this isn’t going to be true and there are certain sequences (highly expressed genes or enriched peaks in a ChIP) where you want the sequence to occur more frequently. Above a certain read density the only way to place more reads into a region is via duplication so we might not expect to have a properly diverse library.
What we can see in this case is that we have lots of sequences with low duplication levels (2-9 copies) which probably cover most of the ‘normal’ genes we’ve measured in the library. Then there are a bunch of sequences with moderate duplication (10 – 100 copies) which may cover things like rRNA and some repeat sequences. There are very few highly duplicated sequences which might be more indicative of a technical problem with the library.
SeqMonk will probably throw a warning or error from these libraries since fairly large chunks of the library will be lost were it to be deduplicated, but the profile seen here actually isn’t too bad for RNA-Seq. If anyone has a suggestion for a better generic metric to use than the amount of loss upon deduplication then we’d be happy to hear suggestions.
Example 3 – PhiX
 In a PhiX control lane you expect that all of the PhiX sequences will be present thousands of times, so we don’t expect this to look pretty. From the blue line we can see that in the raw library the majority of the library is composed of sequences which are present 1000 – 5000 times, with very few sequences from the other categories.
In a PhiX control lane you expect that all of the PhiX sequences will be present thousands of times, so we don’t expect this to look pretty. From the blue line we can see that in the raw library the majority of the library is composed of sequences which are present 1000 – 5000 times, with very few sequences from the other categories.
Interestingly we can also see that when we deduplicate the library that the vast majority of sequences in the deduplicated set would come from reads which were present only once in the original library, suggesting that the deduplicated set would be dominated by sequencing errors or contaminants. This is maybe a good indication of why deduplicating your data as a matter of course is a really bad idea.
Example 4 – Horrible PCR duplication
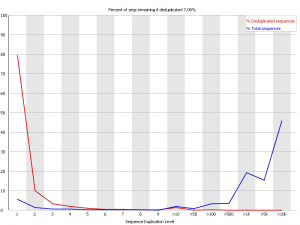 This was about the worst example I could find. This was an RNA-Seq library which had some nasty contamination and PCR duplication. you can see that in the raw data the majority of reads come from sequences which are present tens of thousands of times in the library, but that there is also strong representation from reads which are present hundreds or thousands of times, suggesting that the duplication is widespreads.
This was about the worst example I could find. This was an RNA-Seq library which had some nasty contamination and PCR duplication. you can see that in the raw data the majority of reads come from sequences which are present tens of thousands of times in the library, but that there is also strong representation from reads which are present hundreds or thousands of times, suggesting that the duplication is widespreads.
In this case deduplicating may help the experiment, but it causes the loss of 93% of the original data so it’s a pretty drastic step to take.
3 Comments
Does this version is already available?
If not, what is the ETA for it?
The new version isn’t out yet. We’re still not quite happy with the changes to the Kmer module, but the rest of it is pretty solid. If you want to try a development snapshot then there’s one at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/fastqc_v0.11.0_devel.zip but please be aware this is still a development release so use it at your own risk (but please report any problems you find)
Thanks. I will. Much appreciated.